ViennaRNA 安装使用
RNAfold 是 Vienna RNA Package 里的一种通过计算最小自由能 (MFE: minimum free energy) 来预测 RNA 二级结构的软件。它的算法和理论支持主要来源于 M. Zuker 和 P. Stiegler 在 1981 年发表的这篇 Optimal computer folding of large RNA sequences using thermodynamic and auxiliary information。想了解 Michael Zuker 教授小组的一系列研究情况和 MFE 算法计算依据的可以点这里。
主要参数¶
RNAfold -p -d2 --noLP < sequence1.fa > sequence1.out
详细的参数参考:https://www.tbi.univie.ac.at/RNA/RNAfold.1.html。 本节部分内容来源于:RNAfold 介绍 - 360doc
- -p, –partfunc[=INT] Calculate the partition function and base pairing probability matrix. (default=‘1’)
- -noLP 表示不允许有孤立碱基对;
- -noPS 表示不生成结构图(只计算结果,即不产生二级结构对应的 postscript 文件,这种文件可以转换为 PDF 格式);
- d1 指的是 “dangling end” (悬挂端)的能量计算按照 d1 模式(有且仅有未配对的碱基可以参与最多一个悬挂端),返回值就是“点-弧”结构图和预测的能量值。
关于 dangling end(dangling bases) 的中文翻译在国内未能查到,这里暂翻译为“悬挂端”。可以参考这篇 Long RNA Dangling End Has Large Energetic Contribution to Duplex Stability,dangling end 指的就是在双链中未配对、会参与旁边“链栈”(stacks) 能量增益的碱基。不同的悬挂组合产生的不同的能量增益,这在 RNAfold 计算中也会被考虑(寻求最好的 dangle)。
输出结果¶
默认参数会输出以下两种二级结构:
- optimal secondary structure。最佳二级结构,保证对应的自由能最小,最小自由能简称 MFE,结果示意如下
- centroid secondary structure。自由能表征改变这个结构需要注入的能量大小,对应的数值越小,该结构越稳定。
安装¶
ViennaRNA-2.5.x 要求 C++17 标准的更高版本的编译器,即 GCC 需要 5 及以上版本。
That is unfortunate! We added dlib to our source tree for version 2.5.0 to efficiently compute concentration dependencies upon multi-strand interactions. Apparently, dlib requires a more recent compiler that actually implements the C++17 standard, which is only available since GCC 5. —— From issues 146 我们在版本 2.5.0 中将 dlib 添加到了我们的源代码树中,以便高效地计算多股链交互的浓度依赖性。显然,dlib 需要一个实现了 C++17 标准的更高版本的编译器,这仅在 GCC 5 之后可用。
$ ./configure --prefix=/Bioinfo/Pipeline/SoftWare/ViennaRNA-2.4.11
......
configure:
======================================
ViennaRNA Package 2.4.11
======================================
Sub Packages
------------
* Kinfold : yes
* RNAforester : yes
* Analyse{Dists,Seqs} : no
* RNAlocmin : yes
* Kinwalker : no
Extra Libraries
---------------
* Support Vector Machine : yes
* GNU Scientific Library : no
* GNU MPFR : no
* JSON : yes
Features
--------
* Boustrophedon : yes
* Use hash for NR Sampling : no
* C11 features : yes
* TTY colors : yes
* Float Precision(PF} : no
* Deprecation Warnings : no
Optimizations
-------------
* Auto Vectorization : yes
* Explicit SIMD Extension : yes
* Link Time Optimization : yes
* POSIX Threads : yes
* OpenMP : yes
Scripting Language Interfaces
-----------------------------
* Perl 5 : yes
* Python 2 : yes
* Python 3 : yes
Documentation
-------------
* Reference Manual (PDF) : yes
* Reference Manual (HTML) : yes
* Tutorial (PDF) : yes
* Tutorial (HTML) : no
Unit Tests
----------
* Executable Programs : yes
* C-Library : no
* Perl 5 Interface : yes
* Python 2 Interface : yes
* Python 3 Interface : yes
MacOS X
-------
* Universal Binary : no
* Installer : no
* SDK : custom
Install Directories
-------------------
* Executables : /galaxy-dist/software/ViennaRNA-2.4.11/bin
* Libraries : /galaxy-dist/software/ViennaRNA-2.4.11/lib
* Header files : /galaxy-dist/software/ViennaRNA-2.4.11/include
* Extra Data : /galaxy-dist/software/ViennaRNA-2.4.11/share
* Man pages : /galaxy-dist/software/ViennaRNA-2.4.11/share/man
* Documentation : /galaxy-dist/software/ViennaRNA-2.4.11/share/doc/ViennaRNA
(HTML) : /galaxy-dist/software/ViennaRNA-2.4.11/share/doc/ViennaRNA/html
(PDF) : /galaxy-dist/software/ViennaRNA-2.4.11/share/doc/ViennaRNA
* Perl5 Interface :
(binaries) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/perl5/site_perl/5.26.1/x86_64-linux-thread-multi
(scripts) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/perl5/site_perl/5.26.1
* Python2 Interface :
(binaries) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/python2.7/site-packages
(scripts) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/python2.7/site-packages
* Python3 Interface :
(binaries) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/python3.6/site-packages
(scripts) : /galaxy-dist/software/ViennaRNA-2.4.11/lib/python3.6/site-packages
You can run 'make', 'make check' and 'make install' now!
$ make
$ make install
命令行使用示例。
RNAfold -p -d2 --noLP < sequence1.fa > sequence1.out
使用示例¶
RNAlib Python¶
参考教程:
- https://www.tbi.univie.ac.at/RNA/ViennaRNA/doc/html/helloworld_swig.html
- https://www.tbi.univie.ac.at/RNA/ViennaRNA/doc/html/examples_python.html
import RNA
seq = "GGGCUAUUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGCUGAUUCGAAUUCAGCAUAGCCCA"
md = RNA.md()
# change temperature and dangle model
md.temperature = 20.0 # 20 Deg Celcius
md.dangles = 1 # Dangle Model 1
md.noLP = 1
fc = RNA.fold_compound(seq, md)
(ss, mfe) = fc.mfe()
print("{}\n{} ({:6.2f})".format(seq, ss, mfe))
GGGCUAUUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGCUGAUUCGAAUUCAGCAUAGCCCA
(((((((..((((.........)))).(((((.......))))).....(((((.......)))))))))))). (-28.90)
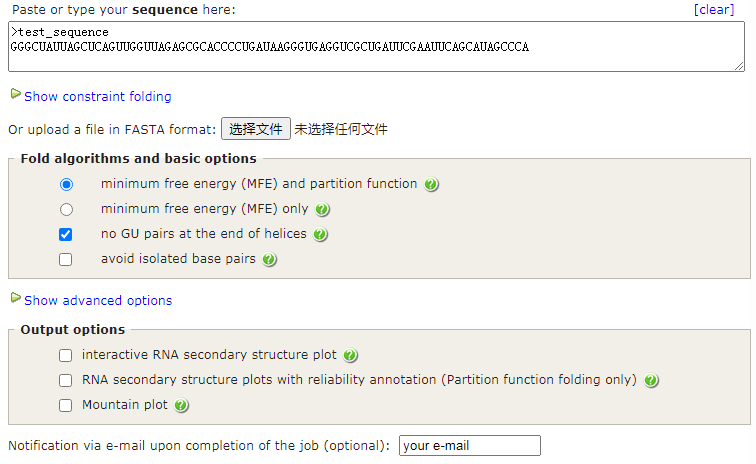
RNAfold web server¶
在线地址:http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi
# Results have been computed using RNAfold 2.4.17. An equivalent command line call would have been
RNAfold -p -d0 --noClosingGU < test_sequenc.fa > test_sequenc.out
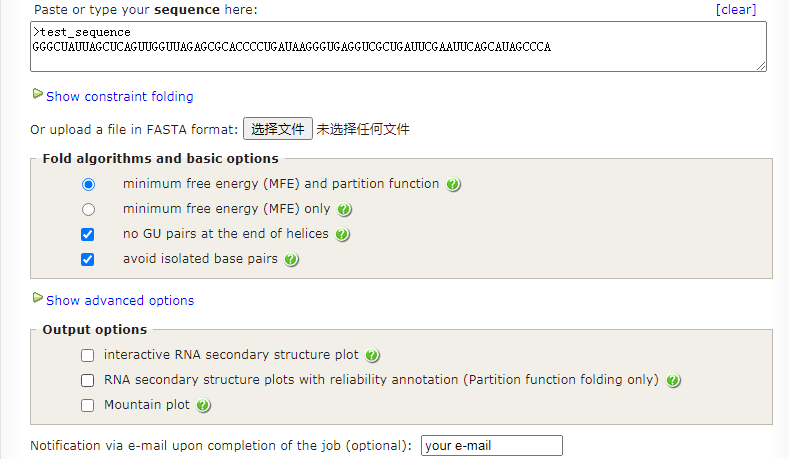
# Results have been computed using RNAfold 2.4.17. An equivalent command line call would have been
RNAfold -p -d0 --noLP --noClosingGU < test_sequenc.fa > test_sequenc.out
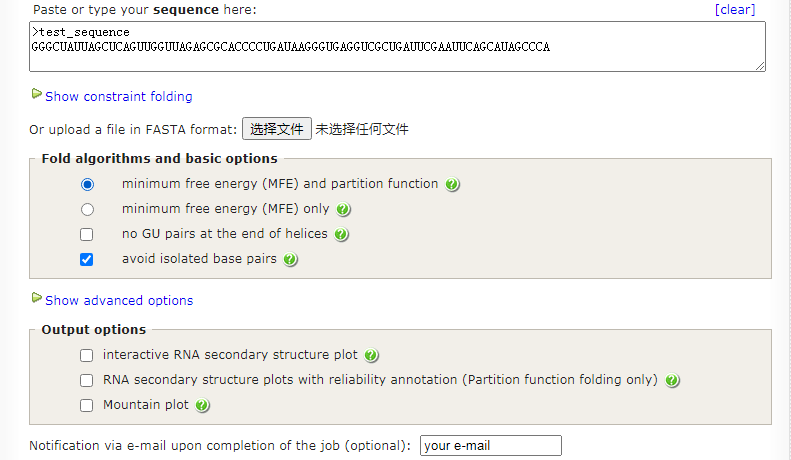
# Results have been computed using RNAfold 2.4.17. An equivalent command line call would have been
RNAfold -p -d0 --noLP < test_sequenc.fa > test_sequenc.out
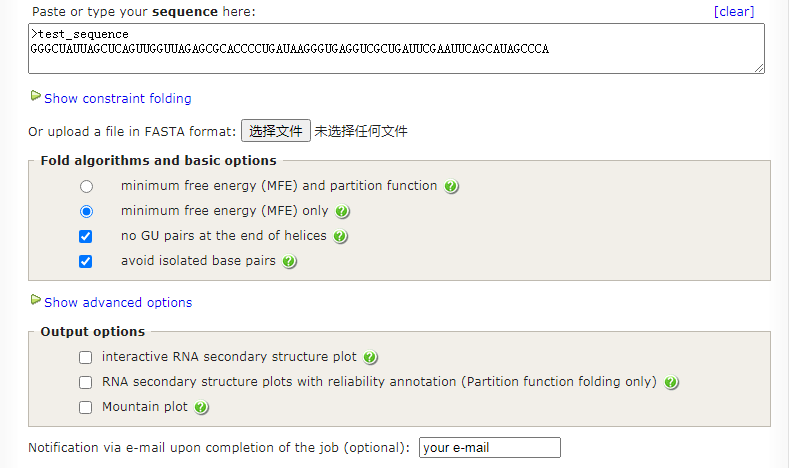
# Results have been computed using RNAfold 2.4.17. An equivalent command line call would have been
RNAfold -d0 --noLP --noClosingGU < test_sequenc.fa > test_sequenc.out
参考资料¶
- TBI - RNAfold - Manpage
- TBI - ViennaRNA Package 2
- Galaxy Tools - vienna_rna
- Galaxy toolshed - viennarna_rnafold